 |
AGA V1
Asexual Genetic Algorithm Version 1.0
|
 |
AGA V1
Asexual Genetic Algorithm Version 1.0
|
module func More...
Functions/Subroutines | |
| subroutine | deltabox (npar, niter, nind, deltaconst, reduc, arrmain, delta0, delta) |
| Updates the size of the hyper-boxes. | |
| real | med (stat, N) |
| Computes the mean value. | |
| real | sigm (stat, N) |
| Computes the standard deviation. | |
| subroutine | spawn (npar, gensize, nmig, ngaussdev, delta, qpar, qbounds, qfam) |
| Asexual reproduction routine. | |
| subroutine | criterion (npar, q, delta, beta, ncrit) |
| Evaluates convergence criterion. | |
| subroutine | randdata (ndata, yobs, ysigma) |
| Generates new realization from experimental data. | |
| real | gaussdev (idum) |
| Generates pseudo-random (Gaussian distributtion) | |
| subroutine | pickbest (npar, nind, nparents, arrmain, fitness, arrpars) |
Select the fittest individuals (nparents number of future parents) from the entire population. Based in the smallest value of the merit function (smaller number=fittest, e.g. 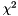 ) ) | |
| subroutine | gaussian (ngaus, i, arrmain) |
module func
This module contains global functions and subroutines used by AGA-V1
| subroutine func::criterion | ( | integer,intent(in) | npar, |
| real,dimension(npar),intent(in) | q, | ||
| real,dimension(npar),intent(in) | delta, | ||
| real,intent(in) | beta, | ||
| integer,intent(out) | ncrit | ||
| ) |
Evaluates convergence criterion.
| [in] | npar | : Number of free parameters |
| [in] | q(npar) | : Individual data |
| [in] | delta(npar) | : Window size of the individual |
| [in] | beta | : Tolerance value |
| [out] | ncrit | : Return value=1 if criterion met, otherwise is not modified |
Definition at line 201 of file modules.f90.
| subroutine func::deltabox | ( | integer,intent(in) | npar, |
| integer,intent(in) | niter, | ||
| integer,intent(in) | nind, | ||
| integer,intent(in) | deltaconst, | ||
| real,intent(in) | reduc, | ||
| real,dimension(npar,nind),intent(in) | arrmain, | ||
| real,dimension(npar),intent(in) | delta0, | ||
| real,dimension(npar),intent(out) | delta | ||
| ) |
Updates the size of the hyper-boxes.
| [in] | npar | : number of free parameters (parameter space dimension, denoted by "n" in the paper) |
| [in] | niter | : number of iteration counter |
| [in] | nind | : number of individials in the entire population |
| [in] | deltaconst | : if equal to 1 the reduction of the hyperboxes is at a constant rate (see reduc), otherwise hyperboxes are reduced with the standard deviation of each parameter. |
| [in] | reduc | : reducing factor, if a constant speed is chosen the hyper-box is reduced according to: 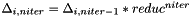 |
| [in] | arrmain(npar,nind) | : Entire population of "nind" individuals, each with "npar" parameters. |
| [in] | delta0(npar) | : Original size of the hyperboxes, array of 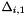 . . |
| [out] | delta(npar) | : array of 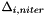 (niter is constant, thus is only a vector w/npar entries). (niter is constant, thus is only a vector w/npar entries). 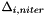 is the size of the hyper-box corresponding to the "i-th" parameter is the size of the hyper-box corresponding to the "i-th" parameter |
Definition at line 75 of file modules.f90.
References sigm().
Referenced by agav1::Algorithm().
| real func::gaussdev | ( | integer | idum | ) |
Generates pseudo-random (Gaussian distributtion)
Modified from "Numerical Recipes in Fortran 77, second ed." (Press et al., Cambridge University Press)
| [in] | idum | : dummy variable (required) |
Definition at line 250 of file modules.f90.
Referenced by spawn().
| subroutine func::gaussian | ( | integer,intent(in) | ngaus, |
| integer,intent(in) | i, | ||
| real,dimension(:,:),intent(inout) | arrmain | ||
| ) |
Definition at line 410 of file modules.f90.
Referenced by agav1::Algorithm().
| real func::med | ( | real,dimension(n),intent(in) | stat, |
| integer,intent(in) | N | ||
| ) |
Computes the mean value.
| [in] | N | : number of data points |
| [in] | stat(N) | : vector from which the mean is to be obtained |
Definition at line 101 of file modules.f90.
Referenced by agav1::Algorithm(), and sigm().
| subroutine func::pickbest | ( | integer,intent(in) | npar, |
| integer,intent(in) | nind, | ||
| integer,intent(in) | nparents, | ||
| real,dimension(npar,nind),intent(in) | arrmain, | ||
| real,dimension(nind),intent(inout) | fitness, | ||
| real,dimension(npar,nparents),intent(out) | arrpars | ||
| ) |
Select the fittest individuals (nparents number of future parents) from the entire population. Based in the smallest value of the merit function (smaller number=fittest, e.g. 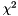 )
)
| [in] | npar | : numbr of free parameters |
| [in] | nind | : number of individuals in the entire population |
| [in] | nparents,: | number of parents, total of fittest individuals to survive for the next generation |
| [in] | arrmain(npar,nind) | : entire population |
| [in,out] | fitness(nind) | : fitness level of each individual, given by the merit function provided b the user in "tarfunc.f90". |
| [out] | arrpars(npar,nparents) | : array of the fittest individuals, which will be parents in the following generation. |
Definition at line 293 of file modules.f90.
References indexx().
Referenced by agav1::Algorithm().
| subroutine func::randdata | ( | integer,intent(in) | ndata, |
| real,dimension(ndata),intent(inout) | yobs, | ||
| real,dimension(ndata),intent(in) | ysigma | ||
| ) |
Generates new realization from experimental data.
| [in] | ndata | : number of experimental data points |
| [in,out] | yobs(ndata) | : experimental data |
| [in] | ysigma(ndata) | : associated uncertainties of yobs |
Definition at line 226 of file modules.f90.
Referenced by agav1::Algorithm().
| real func::sigm | ( | real,dimension(n),intent(in) | stat, |
| integer,intent(in) | N | ||
| ) |
Computes the standard deviation.
| [in] | N | : number of data points |
| [in] | stat(N) | : vector from which the standard deviaiton is to be obtained |
Definition at line 123 of file modules.f90.
References med().
Referenced by agav1::Algorithm(), and deltabox().
| subroutine func::spawn | ( | integer,intent(in) | npar, |
| integer,intent(in) | gensize, | ||
| integer,intent(in) | nmig, | ||
| integer,intent(in) | ngaussdev, | ||
| real,dimension(npar),intent(in) | delta, | ||
| real,dimension(npar),intent(in) | qpar, | ||
| real,dimension(2,npar),intent(in) | qbounds, | ||
| real,dimension(npar,gensize) | qfam | ||
| ) |
Asexual reproduction routine.
| [in] | npar | : number of free parameters |
| [in] | gensize | : size of the new generation from each parent. Since the parent is kept gensize= num of sons + 1 |
| [in] | nmig | : turns on/off migration. If its value is 0, migration is disabled, otherwise is enabled |
| [in] | ngaussdev | : if value equal to 1, offsprinf is obtained with a Gaussian distribution, otherwise they are obtained with uniform random deviates. |
| [in] | delta(npar) | : Size of the parent hyper-box. |
| [in] | qpar(npar) | : parent parameters (its location in the npar-dimensional parameter space |
| [in] | qbounds(npar) | : bounds of the initial universe |
| [out] | qfam(npar,gensize) | : A new family (progeny+parent) |
Definition at line 157 of file modules.f90.
References gaussdev().
Referenced by agav1::Algorithm().
 1.7.4
1.7.4